Database
The Cilento DB is based on the Oracle 10g DB Server on the Entity/Relationship scheme. The scheme is composed of several meta-data areas (pedigrees, phenotypes, genotypes, utilities, general information and laboratory management). The scheme is composed of 37 tables for the first 5 areas, and 79 tables for the last area, where all the history of the study is stored. We have created 53 indexes to increase performance over the E/R model and more than 90 relationships to normalize the database. Security requires us to have different views for each single user, who is authorized to access only specific sections and only in "read mode". Users are able to manage data by themselves with a specific query tool that gives them the possibility to navigate and rebuild a specific result set with minimum knowledge of the system.
Data are stored in three main areas: pedigrees, phenotypes and genotypes.
The pedigrees area contains 31,512 genealogical records. These data have been used to reconstruct very huge pedigrees (15-17 generations) for each population.
Qualitative and quantitative phenotypes, including clinical data, have been collected through physical examination and a structured questionnaire to define personal and family medical history. Biochemical, haematological laboratory tests and instrumental examinations such as ultrasonography (heart, carotids and thyroid), electrocardiogram and bone mineral density tests, have also been carried out. All this information has been collected using a standardized approach through the different populations. Many disease phenotypes and quantitative traits have been checked; particular attention has so far been addressed to cardiovascular traits and diseases.
Genotypes of 1,122 miscrosatellite markers (deCODE map, average marker spacing of 3.6 cM and mean marker heterozygosity of 0.70) for 2,044 individuals have been stored in the genotypes area; SNP genotypes from the 370k Illumina platform are being carried out for 858 individuals. Sequence data of specific gene and mtDNA regions have been generated.
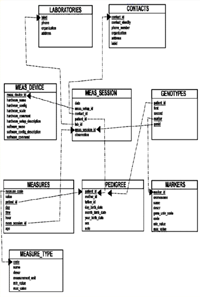
The database does not contain any family and/or first names, all of which information is encrypted by an identification code. An Opteron-based computational cluster has been put in place to distribute efficiently computationally intensive tasks, e.g. QTL analysis, and a job queuing system ensures fair-share policies within the cluster.