Maria R. Matarazzo
Research Director
![]() +39 081 6132426
+39 081 6132426 ![]() maria.matarazzo@igb.cnr.it
maria.matarazzo@igb.cnr.it
Genetics, Genomics and Epigenetics of Diseases
Keywords: Epigenetics, Stem cell identity, DNA methylation, human iPSCs, Primary Immunodeficiencies, Rare disease, CRISPR/Cas9 gene editing
- Research Interest
- Selected Publications
- Professional Experience
- Research Group
Epigenetic control of cellular identity and function in human disorders
In our laboratory we study the role of DNA methylation and the epigenetic modifications in the assembly and maintenance of transcriptional and chromatin domains. How they are perturbed in pathological conditions, such as in primary immunodeficiencies caused by epigenetic regulators (DNMT3B, KMT2D, KDM6A), is the major issue that we are currently addressing. This group of diseases includes ultra-rare and rare genetic disease, like ICF and Kabuki syndromes (KS).
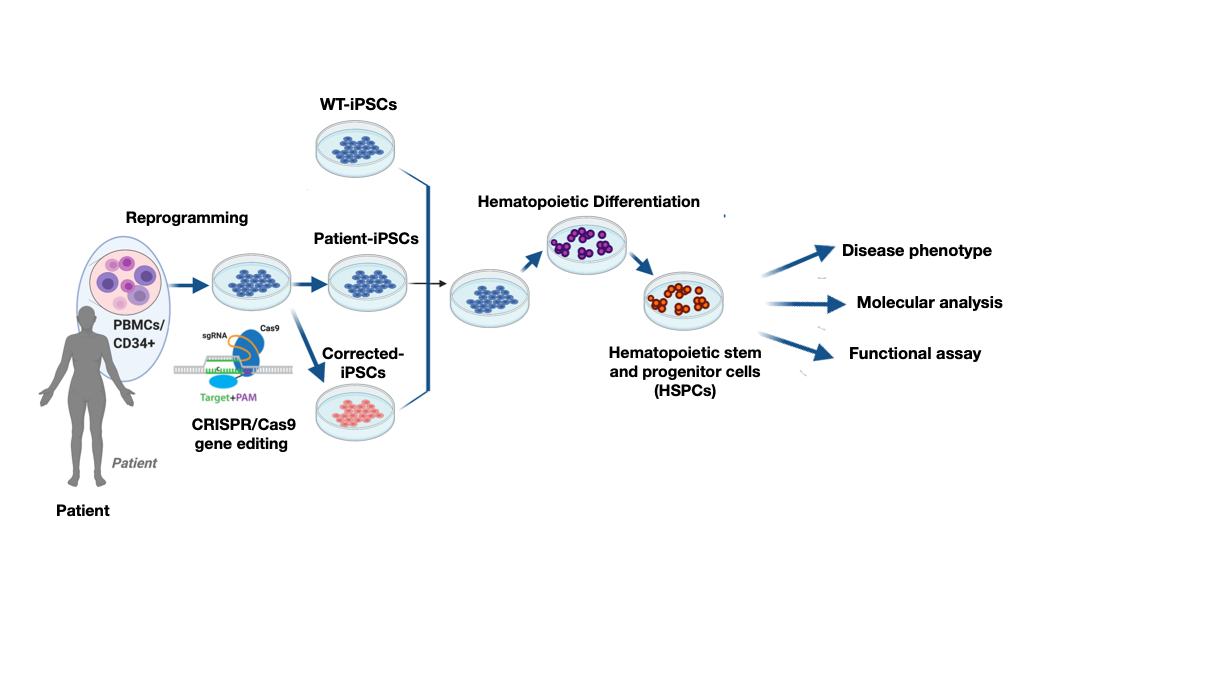
Our experimental strategy combines classic molecular approaches and gene-specific analysis with large-scale transcriptomic and epigenomic technologies in human cellular models, in particular patient-derived induced pluripotent stem cells (iPSCs).
- Development of iPSCs based disease models and CRISPR/Cas9 correction of disease-causing mutations
Because of the lack of proper cellular systems and developmental contexts for molecular studies, nowadays no mechanistic links have been identified between the originally compromised epigenetic function and the cascade of the disease-causing events in primary immunodeficiencies.
Therefore, we contributed to (i) develop a novel disease iPSC-based model for ICF1 syndrome to understand the genotype/phenotype mechanistic links, through recapitulating the process of de novo methylation carried out by DNMT3B during development and (ii) design strategies to restore the proper DNA methylation pattern following CRISPR/Cas9 correction of DNMT3B mutations in ICF1-iPSCs. Currently, we are developing suitable iPSC models to study the early pathogenetic mechanisms of other epigenetic diseases by reprogramming CD34+ cells derived from patients’ peripheral blood.
- Identification of aberrant epigenomes linked to disease cell phenotypes
By comparing the methylation-compromised genome with a corrected ICF genome, we identified genomic targets of de novo methylation mediated by DNMT3B, gaining insights into its deficient activity at disease early stage and into the rescue of its activity. A significant fraction showed a restored methylation pattern, supporting the hypothesis that CRISPR correction of DNMT3B activity might improve the effect of the protein deficiency during later stages of disease development.
Through an integrated multi-omic analysis we detected abnormally increased H3K4me3 levels at genomic regions with persisting DNA hypomethylation in ICF1 iPSC, thus impeding the full DNA methylation rescue. Indeed, pharmacological interference of H3K4me3 deposition improved the DNA methylation recover, thereby alleviating this epigenetic barrier.
Therefore, a strict interconnection among the aberrant epigenetic circuits occurs at the basis of disease cell phenotypes. Hence, reconstructing the defective mechanistic links between DNA and histone methylation marks in patients-iPSCs acquires a valuable significance for their correction.
- Analysis of molecular and cell phenotypes during hematopoietic commitment of patient- and gene corrected- iPSCs
Undifferentiated iPSCs may be in vitro differentiated towards the cell types affected by the disease, thus providing a powerful tool to model and understand disease-related mechanisms. We recently established a differentiation platform of iPSCs towards hematopoietic stem and progenitor cells (HSPCs) in order to study the early immune phenotypes.
1. Fioriniello F, Csukonyi E, Marano D, Brancaccio A, Madonna M, Zarrillo C, Romano A, Marracino F,2 Matarazzo MR, D’Esposito M and Della Ragione F. MeCP2 and major satellite forward RNA cooperate for pericentric heterochromatin organization. Stem Cell Reports. 2020 Dec 8;15(6):1317-1332. 2. Toubiana S, Gagliardi M, Papa M, Manco R, Tzukerman M, Matarazzo MR*, Selig S*. Persistent epigenetic memory impedes rescue of the telomeric phenotype in human ICF iPSCs following DNMT3B correction. Elife. 2019 Nov 20;8:e47859. *Corresponding authors 3. D’Aniello C, Cermola F, Palamidessi A, Wanderlingh LG, Gagliardi M, Migliaccio A, Varrone F, Casalino L, Matarazzo MR, De Cesare D, Scita G, Patriarca EJ, Minchiotti G. Collagen Prolyl Hydroxylation-Dependent Metabolic Perturbation Governs Epigenetic Remodeling and Mesenchymal Transition in Pluripotent and Cancer Cells. Cancer Res. 2019 Jul 1;79(13):3235-3250. 4. Gatto S, Gagliardi M, Franzese M, Leppert S, Papa M, Cammisa M, Grillo G, Velasco G, Francastel C, Toubiana S, D’Esposito M, Angelini C and Matarazzo MR. ICF-specific DNM3B dysfunction interferes with intragenic regulation of mRNA transcription and alternative splicing Nucleic Acids Res. 2017 Jun 2;45(10):5739-5756. 5. Fiorenzano A, Pascale E, Gagliardi M, Terreri S, Papa M, Andolfi G, Galasso M, Tagliazucchi GM, Taccioli C, Patriarca EJ, Cimmino A, Matarazzo MR, Minchiotti G, Fico A. An Ultraconserved Element Containing lncRNA Preserves Transcriptional Dynamics and Maintains ESC Self-Renewal. Stem Cell Reports. 2018 Feb 8. 6. Comes S, Gagliardi M, Laprano N, Fico A, Cimmino A, Palamidessi A, De Cesare D, De Falco S, Angelini C, Scita G, Patriarca EJ, Matarazzo MR* and Minchiotti G*. L-Proline Induces a Mesenchymal-like Invasive Program in Embryonic Stem Cells by Remodeling H3K9 and H3K36 Methylation. Stem Cell Reports, 307-321, 10 October 2013. * Corresponding authors 7. Genesio R, Melis D, Gatto S, Izzo A, Ronga V, Cappuccio G, Lanzo A, Andria G, D’Esposito M, Matarazzo MR*, Conti A*, Nitsch L. Variegated silencing through epigenetic modifications of a large Xq region in a case of balanced X;2 translocation with Incontinentia Pigmenti-like phenotype. Epigenetics. (2011) Oct 1;6(10):1242-7. *Corresponding authors 8. Gatto S, Della Ragione F, Cimmino A, Strazzullo M, Fabbri M, Mutarelli M, Ferraro L, Weisz A, D’Esposito M, Matarazzo MR. Epigenetic alteration of microRNAs in DNMT3B-mutated patients of ICF syndrome. Epigenetics. (2010) Jul 1;5(5):427-43. 9. Talotta F, Cimmino A, Matarazzo MR, De Vita G, D’Esposito M, Di Lauro R, Verde P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene, Jan 8;28(1):73-84 (2009). 10. Matarazzo MR, Boyle S, D’Esposito M, Bickmore WA. (2007). Chromosome territory reorganization in a human disease with altered DNA methylation. Proc Natl Acad Sci U S A 104:16546−16551. 11. Matarazzo MR, De Bonis ML, Strazzullo M, Cerase A, Ferraro M, Vastarelli P, Ballestar E, Esteller M, Kudo S, D’Esposito M. Multiple binding of methyl−CpG and polycomb proteins in long−term gene silencing events. J Cell Physiol. Mar;210(3):711−9 (2007). 12. Fadi J. Charcar, Marta Svartman, Nisrine El−Mogharbel, Mario Ventura, Patrick Kirby, Maria R Matarazzo, Alfredo Ciccodicola, Mariano Rocchi, Maurizio D’Esposito, and Jennifer A. Marshall Graves. Complex Events in the Evolution of the Human Pseudoautosomal Region 2 (PAR2). Genome Research, Feb 13, 281−6 (2003). 13. Matarazzo MR, De Bonis ML, Gregory RI, Vacca M, Hansen RS, Mercadante G, D’Urso M, Feil R, D’Esposito M. Allelic inactivation of the pseudoautosomal gene SYBL1 is controlled by epigenetic mechanisms common to the X and Y chromosomes. Hum Mol Genet., 11, 3191−8. (2002) |
PROFESSIONAL EXPERIENCE AND AWARDS
2010 – Current Researcher II level at IGB-ABT (CNR) and Group Leader of a team studying the role of DNA methylation and the epigenetic modifications in the assembly and maintenance of transcriptional and chromatin domains. How they are perturbed in pathological conditions, such as primary immunodeficiencies caused by epigenetic factors are the major issues that we are currently addressing.
2006 Visiting Researcher at MRC, Human Genetics Unit, Edinburgh, UK. Wendy Bickmore Lab
May-July 2004 CNR Mobility Fellow at MRC, Human Genetics Unit, Edinburgh, UK, to study the nuclear compartmentalization as higher-level control of gene expression and the interplay with others epigenetic marks. Wendy Bickmore Lab
Dec 2001 Researcher III level at IGB-ABT (CNR) studying the role of epigenetic mechanisms in maintenance the X and Y Inactivation of human PAR2 genes.
2000 Telethon Fellow at The Babraham Institute, Cambridge, UK learning methods for studying the remodeling of chromatin structure by histone modifications
1999 EMBO Short Term Fellow at The Babraham Institute, Cambridge, UK. Project title: “Methylation profile and chromatin conformation analysis of the Xq pseudoautosomal gene SYBL1”.
EDUCATION AND TRAINING
2001 PhD in Biochemistry and Molecular Biology University of Naples Federico II.
1996 Degree at University of Naples Federico II cum laude.
![]() +39 081 6132 607
+39 081 6132 607 ![]() maria.strazzullo@igb.cnr.it
maria.strazzullo@igb.cnr.it
Genetics, Genomics and Epigenetics of Diseases
Keywords: DNA methylation, Chromatin biology, human iPSCs, Primary immunodeficiencies, Hematopoiesis, Rare genetic diseases, Cancer epigenetics
Project Title: Molecular and functional characterization of a novel iPSC-based model of ICF2 syndrome